Genomics and bioinformatics: analysis of several large genetic datasets such as the 100,000 genomes project and the uk bio bank.
More than hundred years of research in biology, the main aim to the scientific investigation is the data collection. Later, with the advent of different sources, the aim is moved from data collection to analysis. There are variety of choices available for examining or evaluating an experimental dataset. However, the researchers are still searching for answers for the new biological questions which can be answered by large amount of genomic datasets, molecular data, and model organisms (Welsh et al., 2017).
Analysing large set of genetic data is an upcoming trend in Genomics and Bioinformatics. There are several resources where you can collect large datasets involving biological measurements, lifestyle characteristics, blood details, medical records, etc. In this bog, I will discuss about the Statistical Analysis of large genetic data sets such as 100000 genomes project and the UK biobank.
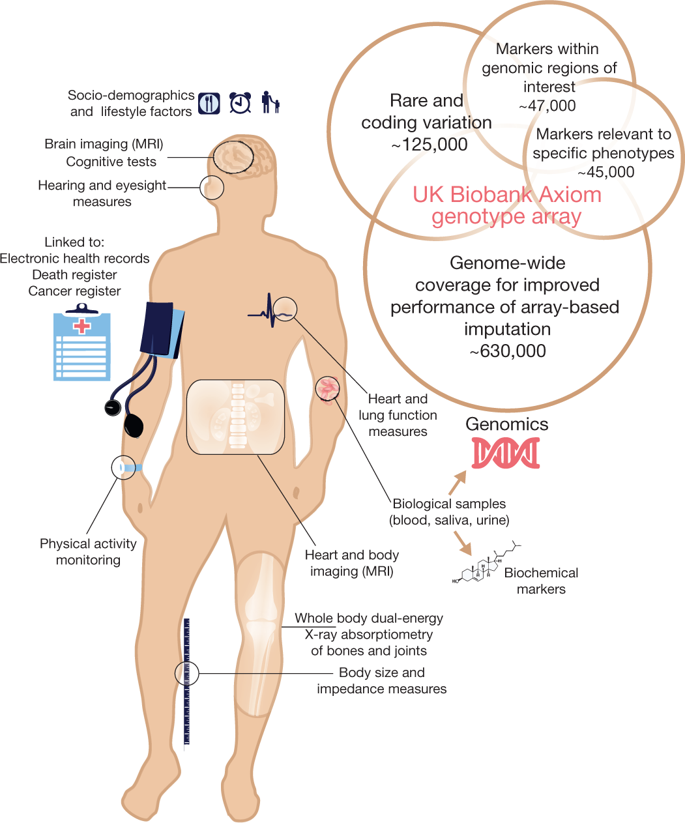
The UK biobank collects a wide variety of phenotypic and biological observations and the data involves the socio-economic factors, health status, physical examination reports, etc. The data also contains the blood samples, urine samples that are stored in proper way to perform different experiments (metabolic analyses). The following figure is the summary of the UK biobank resources and genetic content (Biobank, 2015).
Further, with the huge amount of data, say for example blood samples to test DNA it is not possible to analyse in all samples collected. Thus, sampling methodology is useful for this purpose. Even though, there are many sampling techniques is available in literature, the most widely used for this purpose is the sequential sampling (Manichaikul et al., 2010)
Once the sample of data is collected, it is important to test the quality by conducting the quality control experiments (batch efforts, plate efforts, sex efforts, etc.). Quality check is conducted to avoid misclassification among the samples. In the case of epidemiological analyses, the understanding of close relationship between the person and their siblings is important and these information’s were not collected by the UK bio bank. Depending on the objective of the study, the analysis plan gets modified. Suppose if you want to understand the relationship between the cause and effect of a particular disease, then a simple association test satisfies the purpose. Here is the list of statistical analysis that can suitable for analysing genomic datasets (Bycroft et al., 2018).
- Principal Component analysis (PCA) – Principal component analysis is often called as a dimension reduction technique. If you have hundreds of thousands of samples and PCA will select the most important variables (homogeneous variables) for the study.
- Kinship coefficient estimation – It is the robust estimation method and less time consuming. However, care should be taken in the case of overestimation and underestimation.
- Hidden Markov Model – Hidden Markov Model is used when there is any missing entry in the data.
- Linear Mixed Model – To understand the association between the disease and the other factors causing the disease.
- Bayesian Approach – Bayesian methods or Bayes factor is used to identify the sensitivity and specificity.
- Understanding the pattern in genomics by classification techniques such as regression, Random Forests, decision trees, neural networks, etc. For example, consider the use of statistical method Random Forest to analyse the Hipsterism data. Random Forest model reduce the risk of overfitting compared to other techniques in the machine learning. I have used VariantSpark RF method in scala to analyse the genomic data. Random forest classifies the data by splitting into nodes and group the relevant data into clusters (Cahan et al., 2007).
- Meta Analyses – Used to test the hypothesis for many experimental data.
- Big data analytics – Often Big data Analytics is used in many business problems. However, it is also used in understanding the how genetic research is done. For instance, the National institute of health started adopting the big data analytics to develop genetic guided treatment with proper medicine to improve prevention from disease, early detection and treatment of disease. This can be studied by collecting the health records and are then categorized using Data Mining techniques.
- Machine Learning – ML technique is used for high dimensional datasets to predict the future outcome.
- CRISPR – It is a cluster technology that allows the researchers to add, remove, or alter the genetic material in the DNA. It is cost efficient technique which means it is more affordable even to the public.
In conclusion, with the increase in the technologies like big data, Artificial intelligence, the analysis of genomic data becomes easier than before. Now, there has been a increasing development in the bio-medical industry and there are more concentrating on identifying the disease pattern using Artificial Intelligence (Langmead et al., 2009). Thus, more interesting Statistical Analysis should be adopted for the development of this field.
Above blog referred here
- Biobank, U. Genotyping of 500,000 UK Biobank participants. Description of Sample Processing Workflow and Preparation of DNA for Genotyping, 11, (2015). https://www.ukbiobank.ac.uk/wp-content/uploads/2014/04/UKBiobank_genotyping_QC_documentation-web.pdf
- Bycroft, C., Freeman, C., Petkova, D., Band, G., Elliott, L. T., Sharp, K., Motyer, A., Vukcevic, D., Delaneau, O., & O�Connell, J. The UK Biobank resource with deep phenotyping and genomic data. Nature, 562 7726, (2018), pp. 203–209. https://www.nature.com/articles/s41586-018-0579-z.
- Cahan, P., Rovegno, F., Mooney, D., Newman, J. C., Laurent III, G. S., & McCaffrey, T. A. Meta-analysis of microarray results: challenges, opportunities, and recommendations for standardization. Gene, 401 1–2, (2007), pp. 12–18. https://www.sciencedirect.com/science/article/pii/S0378111907003289
- Langmead, B., Schatz, M. C., Lin, J., Pop, M., & Salzberg, S. L. Searching for SNPs with cloud computing. Genome Biology, 10 11, (2009), pp. R134. https://genomebiology.biomedcentral.com/articles/10.1186/gb-2009-10-11-r134
- Manichaikul, A., Mychaleckyj, J. C., Rich, S. S., Daly, K., Sale, M., & Chen, W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics, 26 22, (2010), pp. 2867–2873. https://academic.oup.com/bioinformatics/article-abstract/26/22/2867/228512
- Welsh, S., Peakman, T., Sheard, S., & Almond, R. Comparison of DNA quantification methodology used in the DNA extraction protocol for the UK Biobank cohort. BMC Genomics, 18 1, (2017), pp. 26. https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-016-3391-x.